The leaves of a plant are often a crucial feature for species identification: the leaf of the Wych Elm (Ulmus glabra) has a serrated edge, the leaves of the Wild Tulip (Tulipa sylvestris) are soft and hairless, and the leaves of the Guelder Rose (Viburnum opulus) are always arranged oppositely. There are many examples of plants whose leaves look so unmistakably that the corresponding species can be confidently identified. However, this does not apply to all plants! Some species respond to environmental influences by changing certain characteristics of their leaf shape, such as the size of the leaf surface or the density of leaf venation.
“Leaf polymorphism” (heterophylly) means that a single plant can have very differently shaped leaves. Some herbs can produce several very different leaf forms, so distinct that they might not be attributed to the same species. A notable example of this is found in some species of the genus Ranunculus, commonly known as buttercups. For instance, the Water Buttercup (genus Ranunculus, section Batrachium) has two completely different leaf forms: the finely divided leaves underwater (submerged leaves) and the roughly three-parted dissected leaves that float (floating leaves).

To understand the phenomenon of heterophylly, one must consider the genetics of the plant. The Water Buttercup has a genotype, a genetic basis for its plant traits, but two different leaf phenotypes (physical appearances). Through years of research, even at the molecular genetic level, scientists have determined which genes need to be activated to initiate the formation of a specific leaf shape. The role of plant growth hormones, their concentrations, and flows that determine the final leaf shape has also been investigated. In the case of the Water Buttercup, the pronounced leaf polymorphism is an adaptation to the environment.
However, there are also examples of leaf polymorphism that remain mysterious to this day, as they cannot be explained by environmental adaptations. Among the early spring plants that push their leaves out of the ground, there is the Goldilocks Buttercup (Ranunculus auricomus agg.). What sets this group of species apart is that the same plant can produce completely different leaf forms, depending on whether it has just emerged from winter dormancy, whether it is flowering, or fruiting.

Within a plant individual, an entire leaf cycle occurs, starting with relatively small and simply three-parted leaves (initial leaves) that emerge immediately after winter. Around April, when the plant starts to bloom, more or less deeply divided leaf forms appear, typically used for species identification. Later in the year, when the plant is fruiting, less dissected leaf forms reappear (summer leaves, final leaves). These resemble the initial leaves but are larger. Why the Goldilocks Buttercup exhibits such leaf diversity within a year remains unclear. An adaptation to the environment, as in the case of the Water Buttercup, could not be demonstrated here.
What remains a puzzle for plant development poses a real challenge for botanical systematists. How can one reasonably morphologically define a plant species like the Goldilocks Buttercup when it constantly changes throughout the year? A tough nut to crack (and a subject of ongoing refinement) even for Flora Incognita!
This article was featured in the Flora-Incognita app as a story in the summer of 2023. The app provides intriguing information about plants, ecology, species identification, as well as tips and tricks for plant identification. Why not take a look?





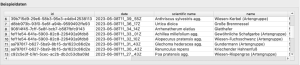
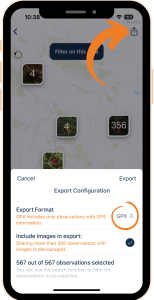 Regardless of the method you choose, first, you need to export your observations from the Flora Incognita app. To do that:
Regardless of the method you choose, first, you need to export your observations from the Flora Incognita app. To do that: